Introduction to reproducible data analysis with R and Quarto
Schedule
Reproducibility and Open Science
- The basics of results reproducibility, replicability, and Open Science
Getting Started with R and Quarto
- Basics of R and Quarto programming
- Integrating code, text, and output within a Quarto document
Working with data
- Creating a data analysis project
- Working with data in R: importing, manipulating, and exporting data
- Data visualization and data analysis
Workshop Requirements
- Are you using the latest version of R?
- Are you on the latest version of RStudio?
👉 Open RStudio
Reproducibility and open science
Reproducibility and replicability
Reproducibility
Reproducibility means that research data and code are made available so that others are able to reach the same results as are claimed in scientific outputs.
Closely related is the concept of replicability, the act of repeating a scientific methodology to reach similar conclusions (using other data).
Reproducibility emphasizes re-using original data and methods for verification, while replicability focuses on achieving consistent results with under similar conditions but using new data.
Open Science for ensuring reproducibility and replicability

The benefits of Open Science for behavioral researchers

Source: The benefits of Open Science
Getting Started with R and Quarto
Change your mental model
Source

Output

Source
---
title: "ggplot2 demo"
author: "Norah Jones"
date: "5/22/2021"
format:
html:
fig-width: 8
fig-height: 4
code-fold: true
---
## Air Quality
@fig-airquality further explores the impact of temperature
on ozone level.
```{r}
#| label: fig-airquality
#| fig-cap: Temperature and ozone level.
#| warning: false
library(ggplot2)
ggplot(airquality, aes(Temp, Ozone)) +
geom_point() +
geom_smooth(method = "loess"
)
```Output 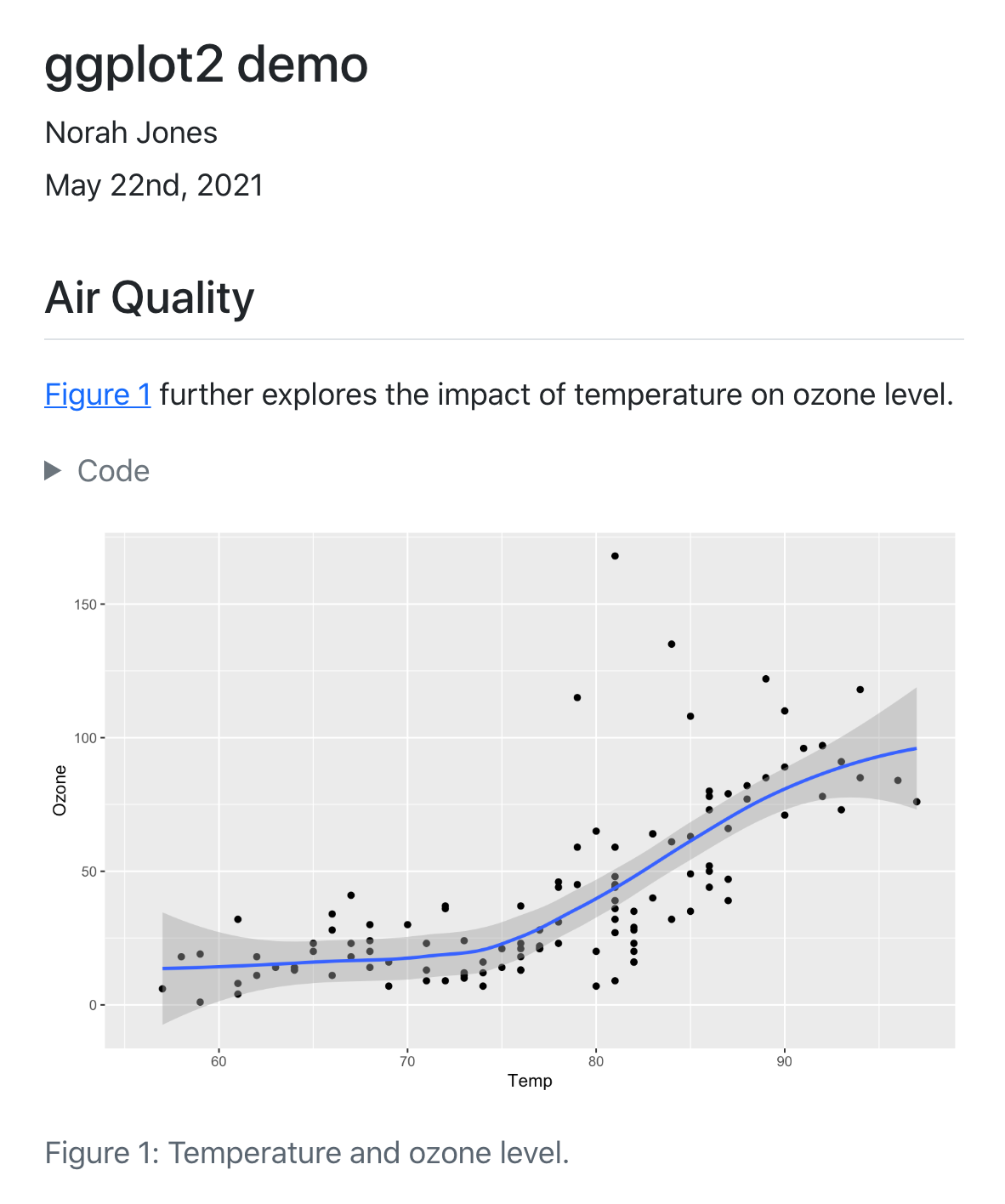
Working with R and RStudio
Getting Started with R and RStudio

Create a folder for your R project

Create an R project

R projects
When a project is opened within RStudio the following actions are taken:
🦋 A new R session is started
🐛 The .Rprofile, .RData file, and .Rhistory files in the project’s main directory are loaded
🐝 The current working directory is set to the project directory
🐞 Other RStudio settings (e.g. active tabs, splitter positions, etc.) are restored to where they were the last time the project was closed
Create a Quarto document (report.qmd)

Install some packages
Quarto
How does Quarto work?

So what is Quarto?
Quarto is a command line interface (CLI) that renders plain text formats (
.qmd,.rmd,.md) into static PDF/Word/HTML reports, books, websites, presentations and more
Rendering
For rendering to pdfs see 👉 here
For rendering to word docs see 👉 here
Working with a .qmd
A .qmd is a plain text file
Metadata: YAML
The YAML header:
influences the final document in different ways. It is placed at the very beginning of the document. The information that it contains can affect the code, content, and the rendering process.
YAML
See more formats and other YAML metadata options here
Markdown
Quarto uses markdown as its underlying document syntax. Markdown is a plain text format that is designed to be easy to write, and, even more importantly, easy to read
Text Formatting
| Markdown Syntax | Output |
|---|---|
|
italics and bold |
|
superscript2 / subscript2 |
|
|
|
verbatim code |
Headings
| Markdown Syntax | Output |
|---|---|
|
Header 1 |
|
Header 2 |
|
Header 3 |
|
Header 4 |
Code
Working with data
Get data
- Download the example dataset
- Store data file in “data/raw” folder
Reading data in
Manipulating data
```{r}
# Name correction
names(data) <- gsub("d2priv", "dapriv2", names(data))
# Own function
recode_5 <- function(x) {
x * (-1)+6
}
# Use of mutate_at to apply function
data_proc <- data %>%
mutate_at(vars(matches("gattAI1_3|gattAI1_6|gattAI1_8|gattAI1_9|gattAI1_10|gattAI2_5|gattAI2_9|gattAI2_10")), recode_5)
```Manipulating data: Creating composites
```{r}
data <- data[ , purrr::map_lgl(data, is.numeric)] %>% # select numeric variables
select(matches("gattAI1|soctechblind|trust1|anxty1|SocInf1|Age")) # select relevant variables
comp_split <- data %>% sjlabelled::remove_all_labels(.) %>%
split.default(sub("_.*", "", names(data))) # creating a list of dataframes, where each dataframe consists of the columns from the original data that shared the same prefix (all characters before the underscore)
comp <- purrr::map(comp_split, ~ rowMeans(.x, na.rm=T)) #calculating the row-wise mean of each data frame in the list `comp_split`, with the output being a new list (`comp`) where each element is a numeric vector of row means from each corresponding data frame in `comp_split`
comp_df <- do.call("cbind", comp) %>% as.data.frame(.) # binding all the elements in the list `comp` into a single data frame, `comp_df`
```Exporting data
The R folder
Storing and sourcing custom functions from the R folder
- You may store your custom functions in a separate file (e.g., “R/custom-functions.R”) that you may source in your report document (“report.qmd”)
In R/custom-functions.R:
In report.qmd:
Visualisations
Reading in pre-processed data
Creating a correlation plot

Scatterplot

Saving scatterplot
- Save your scatterplot in an output/figs folder
Review your folder structure
Questions? Remarks?
takk for oppmerksomheten!


